Pre-trained ODAC models are versatile across various MOF-related tasks. To begin, we’ll start with a fundamental application: calculating the adsorption energy for a single CO2 molecule. This serves as an excellent and simple demonstration of what you can achieve with these datasets and models.
For predicting the adsorption energy of a single CO2 molecule within a MOF structure, the adsorption energy () is defined as:
Each term on the right-hand side represents the energy of the relaxed state of the indicated chemical system. For a comprehensive understanding of our methodology for computing these adsorption energies, please refer to our paper.
Loading Pre-trained Models¶
Need to install fairchem-core or get UMA access or getting permissions/401 errors?
Install the necessary packages using pip, uv etc
! pip install fairchem-core fairchem-data-oc fairchem-applications-cattsunamiGet access to any necessary huggingface gated models
Get and login to your Huggingface account
Request access to https://
huggingface .co /facebook /UMA Create a Huggingface token at https://
huggingface .co /settings /tokens/ with the permission “Permissions: Read access to contents of all public gated repos you can access” Add the token as an environment variable using
huggingface-cli loginor by setting the HF_TOKEN environment variable.
# Login using the huggingface-cli utility
! huggingface-cli login
# alternatively,
import os
os.environ['HF_TOKEN'] = 'MY_TOKEN'A pre-trained model can be loaded using FAIRChemCalculator. In this example, we’ll employ UMA to determine the CO2 adsorption energies.
from fairchem.core import FAIRChemCalculator, pretrained_mlip
predictor = pretrained_mlip.get_predict_unit("uma-s-1p1")
calc = FAIRChemCalculator(predictor, task_name="odac")WARNING:root:device was not explicitly set, using device='cuda'.
WARNING:root:If 'dataset_list' is provided in the config, the code assumes that each dataset maps to itself. Please use 'dataset_mapping' as 'dataset_list' is deprecated and will be removed in the future.
Adsorption in rigid MOFs: CO2 Adsorption Energy in Mg-MOF-74¶
Let’s apply our knowledge to Mg-MOF-74, a widely studied MOF known for its excellent CO2 adsorption properties. Its structure comprises magnesium atomic complexes connected by a carboxylated and oxidized benzene ring, serving as an organic linker. Previous studies consistently report the CO2 adsorption energy for Mg-MOF-74 to be around -0.40 eV [1] [2] [3].
Our goal is to verify if we can achieve a similar value by performing a simple single-point calculation using UMA. In the ODAC23 dataset, all MOF structures are identified by their CSD (Cambridge Structural Database) code. For Mg-MOF-74, this code is OPAGIX. We’ve extracted a specific OPAGIX+CO2 configuration from the dataset, which exhibits the lowest adsorption energy among its counterparts.
import matplotlib.pyplot as plt
from ase.io import read
from ase.visualize.plot import plot_atoms
mof_co2 = read("structures/OPAGIX_w_CO2.cif")
mof = read("structures/OPAGIX.cif")
co2 = read("structures/co2.xyz")
fig, ax = plt.subplots(figsize=(5, 4.5), dpi=250)
plot_atoms(mof_co2, ax)
ax.set_axis_off()
The final step in calculating the adsorption energy involves connecting the FAIRChemCalculator to each relaxed structure: OPAGIX+CO2, OPAGIX, and CO2. The structures used here are already relaxed from ODAC23. For simplicity, we assume here that further relaxations can be neglected. We will show how to go beyond this assumption in the next section.
mof_co2.calc = calc
mof.calc = calc
co2.calc = calc
E_ads = (
mof_co2.get_potential_energy()
- mof.get_potential_energy()
- co2.get_potential_energy()
)
print(f"Adsorption energy of CO2 in Mg-MOF-74: {E_ads:.3f} eV")Adsorption energy of CO2 in Mg-MOF-74: -0.459 eV
Adsorption in flexible MOFs¶
The adsorption energy calculation method outlined above is typically performed with rigid MOFs for simplicity. Both experimental and modeling literature have shown, however, that MOF flexibility can be important in accurately capturing the underlying chemistry of adsorption [1] [2] [3]. In particular, uptake can be improved by treating MOFs as flexible. Two types of MOF flexibility can be considered: intrinsic flexibility and deformation induced by guest molecules. In the Open DAC Project, we consider the latter MOF deformation by allowing the atomic positions of the MOF to relax during geometry optimization [4]. The addition of additional degrees of freedoms can complicate the computation of the adsorption energy and necessitates an extra step in the calculation procedure.
The figure below shows water adsorption in the MOF with CSD code WOBHEB with added defects (WOBHEB_0.11_0) from a DFT simulation. A typical adsorption energy calculation would only seek to capture the effects shaded in purple, which include both chemisorption and non-bonded interactions between the host and guest molecule. When allowing the MOF to relax, however, the adsorption energy also includes the energetic effect of the MOF deformation highlighted in green.
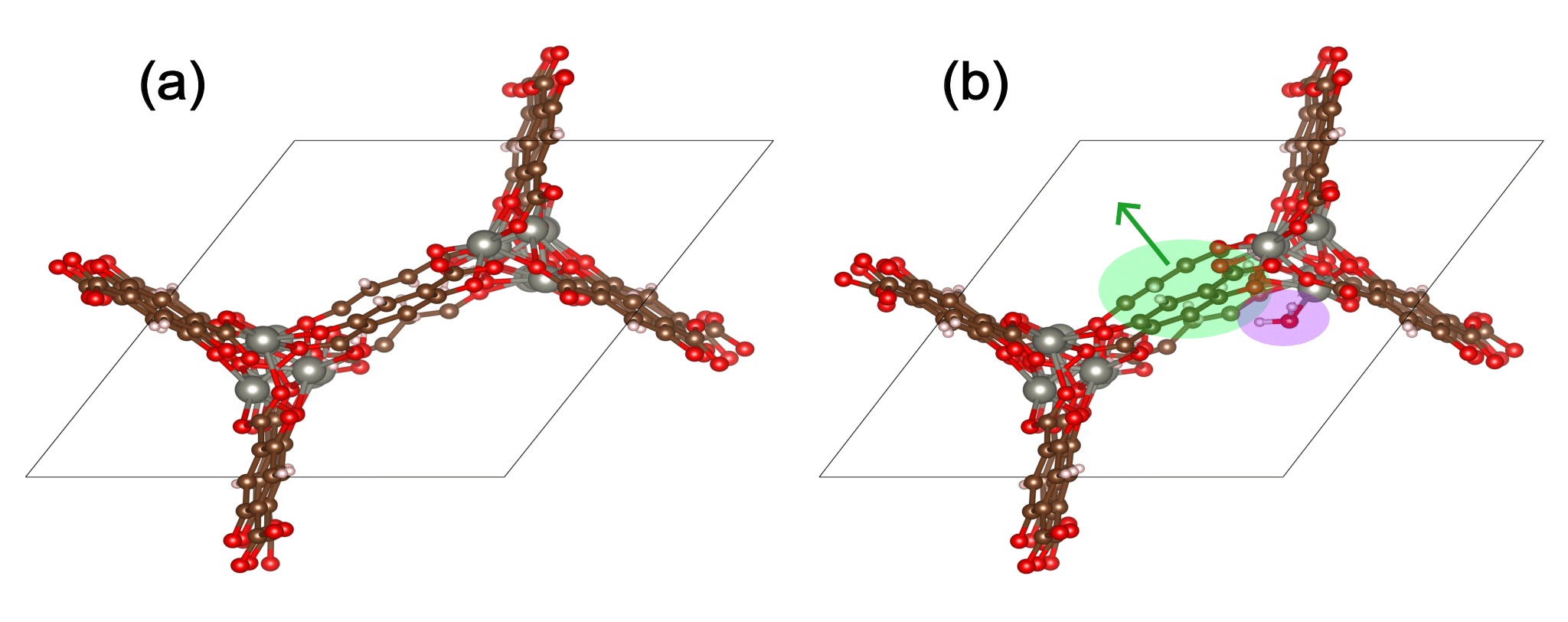
To account for this deformation, it is vital to use the most energetically favorable MOF geometry for the empty MOF term in Eqn. 1. Including MOF atomic coordinates as degrees of freedom can result in three possible outcomes:
The MOF does not deform, so the energies of the relaxed empty MOF and the MOF in the adsorbed state are the same
The MOF deforms to a less energetically favorable geometry than its ground state
The MOF locates a new energetically favorable geoemtry relative to the empty MOF relaxation
The first outcome requires no additional computation because the MOF rigidity assumption is valid. The second outcome represents physical and reversible deformation where the MOF returns to its empty ground state upon removal of the guest molecule. The third outcome is often the result of the guest molecule breaking local symmetry. We also found cases in ODAC in which both outcomes 2 and 3 occur within the same MOF.
To ensure the most energetically favorable empty MOF geometry is found, an addition empty MOF relaxation should be performed after MOF + adsorbate relaxation. The guest molecule should be removed, and the MOF should be relaxed starting from its geometry in the adsorbed state. If all deformation is reversible, the MOF will return to its original empty geometry. Otherwise, the lowest energy (most favorable) MOF geometry should be taken as the reference energy, , in Eqn. 1.
H2O Adsorption Energy in Flexible WOBHEB with UMA¶
The first part of this tutorial demonstrates how to perform a single point adsorption energy calculation using UMA. To treat MOFs as flexible, we perform all calculations on geometries determined by geometry optimization. The following example corresponds to the figure shown above (H2O adsorption in WOBHEB_0.11_0).
In this tutorial, corresponds to the energy of determined from geometry optimization of .
First, we obtain the energy of the empty MOF from relaxation of only the MOF:
import ase.io
from ase.optimize import BFGS
mof = ase.io.read("structures/WOBHEB_0.11.cif")
mof.calc = calc
relax = BFGS(mof)
relax.run(fmax=0.05)
E_mof_empty = mof.get_potential_energy()
print(f"Energy of empty MOF: {E_mof_empty:.3f} eV") Step Time Energy fmax
BFGS: 0 23:05:08 -1077.274065 0.206406
BFGS: 1 23:05:09 -1077.276780 0.152729
BFGS: 2 23:05:10 -1077.281942 0.169926
BFGS: 3 23:05:11 -1077.284771 0.155763
BFGS: 4 23:05:11 -1077.288834 0.108771
BFGS: 5 23:05:11 -1077.291023 0.086446
BFGS: 6 23:05:12 -1077.293362 0.093448
BFGS: 7 23:05:13 -1077.295414 0.100138
BFGS: 8 23:05:13 -1077.297829 0.102535
BFGS: 9 23:05:15 -1077.300013 0.091603
BFGS: 10 23:05:17 -1077.302011 0.078958
BFGS: 11 23:05:17 -1077.304137 0.105563
BFGS: 12 23:05:17 -1077.306724 0.087907
BFGS: 13 23:05:18 -1077.309517 0.086364
BFGS: 14 23:05:18 -1077.312263 0.086863
BFGS: 15 23:05:19 -1077.314701 0.106278
BFGS: 16 23:05:19 -1077.316986 0.106217
BFGS: 17 23:05:20 -1077.319484 0.085427
BFGS: 18 23:05:20 -1077.322263 0.109640
BFGS: 19 23:05:21 -1077.325135 0.148669
BFGS: 20 23:05:21 -1077.327766 0.125926
BFGS: 21 23:05:23 -1077.329920 0.069120
BFGS: 22 23:05:25 -1077.331952 0.087277
BFGS: 23 23:05:26 -1077.334273 0.125237
BFGS: 24 23:05:27 -1077.336835 0.166696
BFGS: 25 23:05:27 -1077.339543 0.145540
BFGS: 26 23:05:28 -1077.342148 0.087640
BFGS: 27 23:05:28 -1077.344542 0.076211
BFGS: 28 23:05:29 -1077.346893 0.148940
BFGS: 29 23:05:29 -1077.349784 0.170253
BFGS: 30 23:05:29 -1077.352529 0.109272
BFGS: 31 23:05:30 -1077.354751 0.070349
BFGS: 32 23:05:30 -1077.356778 0.089705
BFGS: 33 23:05:30 -1077.358657 0.124280
BFGS: 34 23:05:31 -1077.360597 0.108072
BFGS: 35 23:05:32 -1077.362471 0.068554
BFGS: 36 23:05:32 -1077.364166 0.070187
BFGS: 37 23:05:32 -1077.365712 0.105459
BFGS: 38 23:05:33 -1077.367257 0.104161
BFGS: 39 23:05:33 -1077.368767 0.063019
BFGS: 40 23:05:36 -1077.370140 0.058367
BFGS: 41 23:05:39 -1077.371383 0.058770
BFGS: 42 23:05:41 -1077.372482 0.063346
BFGS: 43 23:05:41 -1077.373506 0.055382
BFGS: 44 23:05:42 -1077.374498 0.042291
Energy of empty MOF: -1077.374 eV
Next, we add the H2O guest molecule and relax the MOF + adsorbate to obtain .
mof_h2o = ase.io.read("structures/WOBHEB_H2O.cif")
mof_h2o.calc = calc
relax = BFGS(mof_h2o)
relax.run(fmax=0.05)
E_combo = mof_h2o.get_potential_energy()
print(f"Energy of MOF + H2O: {E_combo:.3f} eV") Step Time Energy fmax
BFGS: 0 23:05:42 -1091.565588 1.145036
BFGS: 1 23:05:43 -1091.585064 0.314149
BFGS: 2 23:05:43 -1091.590212 0.243429
BFGS: 3 23:05:43 -1091.608167 0.237259
BFGS: 4 23:05:44 -1091.614633 0.227909
BFGS: 5 23:05:44 -1091.625216 0.186828
BFGS: 6 23:05:45 -1091.632353 0.178927
BFGS: 7 23:05:47 -1091.640624 0.175072
BFGS: 8 23:05:47 -1091.648039 0.184491
BFGS: 9 23:05:47 -1091.656144 0.160958
BFGS: 10 23:05:48 -1091.663841 0.178406
BFGS: 11 23:05:48 -1091.672297 0.188726
BFGS: 12 23:05:48 -1091.682087 0.157487
BFGS: 13 23:05:49 -1091.692978 0.177282
BFGS: 14 23:05:49 -1091.704433 0.158175
BFGS: 15 23:05:50 -1091.715510 0.191790
BFGS: 16 23:05:50 -1091.725708 0.197778
BFGS: 17 23:05:50 -1091.735325 0.163735
BFGS: 18 23:05:51 -1091.745537 0.151535
BFGS: 19 23:05:51 -1091.754020 0.170889
BFGS: 20 23:05:51 -1091.761492 0.153765
BFGS: 21 23:05:51 -1091.767890 0.152302
BFGS: 22 23:05:53 -1091.774202 0.165875
BFGS: 23 23:05:54 -1091.780888 0.135516
BFGS: 24 23:05:54 -1091.788351 0.181080
BFGS: 25 23:05:54 -1091.794280 0.204584
BFGS: 26 23:05:55 -1091.800636 0.131359
BFGS: 27 23:05:55 -1091.806517 0.189556
BFGS: 28 23:05:55 -1091.812306 0.199112
BFGS: 29 23:05:56 -1091.817179 0.151644
BFGS: 30 23:05:56 -1091.822212 0.100162
BFGS: 31 23:05:56 -1091.826296 0.125746
BFGS: 32 23:05:58 -1091.832529 0.177403
BFGS: 33 23:06:00 -1091.837115 0.246717
BFGS: 34 23:06:01 -1091.842047 0.112437
BFGS: 35 23:06:01 -1091.845780 0.328522
BFGS: 36 23:06:03 -1091.850877 0.173979
BFGS: 37 23:06:04 -1091.858477 0.159966
BFGS: 38 23:06:05 -1091.865287 0.141074
BFGS: 39 23:06:05 -1091.872005 0.142507
BFGS: 40 23:06:05 -1091.878354 0.242183
BFGS: 41 23:06:06 -1091.881268 0.524486
BFGS: 42 23:06:06 -1091.884787 0.733810
BFGS: 43 23:06:06 -1091.893713 0.205201
BFGS: 44 23:06:07 -1091.898874 0.154866
BFGS: 45 23:06:07 -1091.917403 0.317381
BFGS: 46 23:06:09 -1091.925001 0.327346
BFGS: 47 23:06:09 -1091.943632 0.232350
BFGS: 48 23:06:10 -1091.959187 0.458501
BFGS: 49 23:06:10 -1091.965186 1.155130
BFGS: 50 23:06:11 -1091.964980 1.463737
BFGS: 51 23:06:11 -1092.005735 0.559884
BFGS: 52 23:06:12 -1092.021118 0.289973
BFGS: 53 23:06:12 -1092.063491 0.335282
BFGS: 54 23:06:13 -1092.082404 0.325419
BFGS: 55 23:06:13 -1092.104882 0.607129
BFGS: 56 23:06:15 -1092.121690 0.229615
BFGS: 57 23:06:16 -1092.138181 0.229395
BFGS: 58 23:06:16 -1092.150540 0.188631
BFGS: 59 23:06:16 -1092.164646 0.276043
BFGS: 60 23:06:16 -1092.177335 0.433006
BFGS: 61 23:06:17 -1092.188076 0.446372
BFGS: 62 23:06:17 -1092.198851 0.340839
BFGS: 63 23:06:17 -1092.209976 0.189507
BFGS: 64 23:06:18 -1092.221014 0.140018
BFGS: 65 23:06:18 -1092.229585 0.155958
BFGS: 66 23:06:18 -1092.235876 0.120796
BFGS: 67 23:06:19 -1092.241382 0.097202
BFGS: 68 23:06:19 -1092.247013 0.124405
BFGS: 69 23:06:20 -1092.252422 0.168117
BFGS: 70 23:06:20 -1092.257364 0.155581
BFGS: 71 23:06:20 -1092.262424 0.123190
BFGS: 72 23:06:21 -1092.266808 0.153426
BFGS: 73 23:06:22 -1092.270517 0.176623
BFGS: 74 23:06:23 -1092.274072 0.168299
BFGS: 75 23:06:24 -1092.277533 0.117553
BFGS: 76 23:06:24 -1092.280814 0.081266
BFGS: 77 23:06:25 -1092.284107 0.088314
BFGS: 78 23:06:27 -1092.286626 0.105022
BFGS: 79 23:06:28 -1092.289067 0.090483
BFGS: 80 23:06:28 -1092.291300 0.065899
BFGS: 81 23:06:29 -1092.293091 0.076137
BFGS: 82 23:06:31 -1092.294691 0.080392
BFGS: 83 23:06:31 -1092.296042 0.094850
BFGS: 84 23:06:32 -1092.297503 0.090991
BFGS: 85 23:06:32 -1092.298942 0.097425
BFGS: 86 23:06:32 -1092.300668 0.084134
BFGS: 87 23:06:33 -1092.302339 0.063826
BFGS: 88 23:06:33 -1092.304097 0.053615
BFGS: 89 23:06:33 -1092.305796 0.055280
BFGS: 90 23:06:35 -1092.307151 0.054058
BFGS: 91 23:06:35 -1092.308452 0.048034
Energy of MOF + H2O: -1092.308 eV
We can now isolate the MOF atoms from the relaxed MOF + H2O geometry and see that the MOF has adopted a geometry that is less energetically favorable than the empty MOF by ~0.2 eV. The energy of the MOF in the adsorbed state corresponds to .
mof_adsorbed_state = mof_h2o[:-3]
mof_adsorbed_state.calc = calc
E_mof_adsorbed_state = mof_adsorbed_state.get_potential_energy()
print(f"Energy of MOF in the adsorbed state: {E_mof_adsorbed_state:.3f} eV")Energy of MOF in the adsorbed state: -1077.091 eV
H2O adsorption in this MOF appears to correspond to Case #2 as outlined above. We can now perform re-relaxation of the empty MOF starting from the geometry.
relax = BFGS(mof_adsorbed_state)
relax.run(fmax=0.05)
E_mof_rerelax = mof_adsorbed_state.get_potential_energy()
print(f"Energy of re-relaxed empty MOF: {E_mof_rerelax:.3f} eV") Step Time Energy fmax
BFGS: 0 23:06:36 -1077.091356 0.982171
BFGS: 1 23:06:36 -1077.123575 0.870930
BFGS: 2 23:06:36 -1077.172816 0.806666
BFGS: 3 23:06:37 -1077.212851 0.502350
BFGS: 4 23:06:37 -1077.232427 0.440263
BFGS: 5 23:06:39 -1077.249954 0.284640
BFGS: 6 23:06:39 -1077.260700 0.260798
BFGS: 7 23:06:40 -1077.270187 0.247485
BFGS: 8 23:06:40 -1077.279525 0.211762
BFGS: 9 23:06:41 -1077.286456 0.178535
BFGS: 10 23:06:41 -1077.291571 0.149031
BFGS: 11 23:06:43 -1077.295635 0.139213
BFGS: 12 23:06:43 -1077.299260 0.145840
BFGS: 13 23:06:44 -1077.303894 0.182732
BFGS: 14 23:06:46 -1077.308574 0.171438
BFGS: 15 23:06:46 -1077.313120 0.134348
BFGS: 16 23:06:47 -1077.317226 0.165142
BFGS: 17 23:06:47 -1077.321450 0.150408
BFGS: 18 23:06:48 -1077.325841 0.145401
BFGS: 19 23:06:48 -1077.329411 0.114699
BFGS: 20 23:06:48 -1077.332035 0.108752
BFGS: 21 23:06:48 -1077.334287 0.101454
BFGS: 22 23:06:50 -1077.336599 0.123644
BFGS: 23 23:06:51 -1077.339326 0.119983
BFGS: 24 23:06:53 -1077.341968 0.088932
BFGS: 25 23:06:53 -1077.344302 0.091589
BFGS: 26 23:06:55 -1077.346225 0.068003
BFGS: 27 23:06:55 -1077.347774 0.068608
BFGS: 28 23:06:58 -1077.349486 0.090820
BFGS: 29 23:07:01 -1077.351045 0.096652
BFGS: 30 23:07:05 -1077.352657 0.063820
BFGS: 31 23:07:05 -1077.353839 0.050151
BFGS: 32 23:07:05 -1077.355144 0.054063
BFGS: 33 23:07:06 -1077.356513 0.085249
BFGS: 34 23:07:06 -1077.357918 0.079802
BFGS: 35 23:07:06 -1077.359456 0.069575
BFGS: 36 23:07:07 -1077.360867 0.071534
BFGS: 37 23:07:07 -1077.362149 0.073259
BFGS: 38 23:07:08 -1077.363339 0.084855
BFGS: 39 23:07:08 -1077.364672 0.080591
BFGS: 40 23:07:09 -1077.365666 0.067641
BFGS: 41 23:07:11 -1077.366698 0.058106
BFGS: 42 23:07:11 -1077.367796 0.060969
BFGS: 43 23:07:11 -1077.368842 0.069386
BFGS: 44 23:07:12 -1077.370096 0.076307
BFGS: 45 23:07:12 -1077.371267 0.048851
Energy of re-relaxed empty MOF: -1077.371 eV
The MOF returns to its original empty reference energy upon re-relaxation, confirming that this deformation is physically relevant and is induced by the adsorbate molecule. In Case #3, this re-relaxed energy will be more negative (more favorable) than the original empty MOF relaxation. Thus, we take the reference empty MOF energy ( in Eqn. 1) to be the minimum of the original empty MOF energy and the re-relaxed MOf energy:
E_mof = min(E_mof_empty, E_mof_rerelax)
# get adsorbate reference energy
h2o = mof_h2o[-3:]
h2o.calc = calc
E_h2o = h2o.get_potential_energy()
# compute adsorption energy
E_ads = E_combo - E_mof - E_h2o
print(f"Adsorption energy of H2O in WOBHEB_0.11_0: {E_ads:.3f} eV")Adsorption energy of H2O in WOBHEB_0.11_0: -0.686 eV
This adsorption energy closely matches that from DFT (–0.699 eV) [1]. The strong adsorption energy is a consequence of both H2O chemisorption and MOF deformation. We can decompose the adsorption energy into contributions from these two factors. Assuming rigid H2O molecules, we define and , respectively, as
describes host host–guest interactions for the MOF in the adsorbed state only. quantifies the magnitude of deformation between the MOF in the adsorbed state and the most energetically favorable empty MOF geometry determined from the workflow presented here. It can be shown that
For H2O adsorption in WOBHEB_0.11, we have
E_int = E_combo - E_mof_adsorbed_state - E_h2o
print(f"E_int: {E_int}")E_int: -0.9692270755765957
E_mof_deform = E_mof_adsorbed_state - E_mof_empty
print(f"E_mof_deform: {E_mof_deform}")E_mof_deform: 0.2831425666809082
E_ads = E_int + E_mof_deform
print(f"E_ads: {E_ads}")E_ads: -0.6860845088956875
is equivalent to when the MOF is assumed to be rigid. In this case, failure to consider adsorbate-induced deformation would result in an overestimation of the adsorption energy magnitude.
Acknowledgements & Authors¶
Logan Brabson and Sihoon Choi (Georgia Tech) and the OpenDAC project.
- Sriram, A., Choi, S., Yu, X., Brabson, L. M., Das, A., Ulissi, Z., Uyttendaele, M., Medford, A. J., & Sholl, D. S. (2024). The Open DAC 2023 Dataset and Challenges for Sorbent Discovery in Direct Air Capture. ACS Central Science, 10(5), 923–941. 10.1021/acscentsci.3c01629
- Queen, W. L., Hudson, M. R., Bloch, E. D., Mason, J. A., Gonzalez, M. I., Lee, J. S., Gygi, D., Howe, J. D., Lee, K., Darwish, T. A., James, M., Peterson, V. K., Teat, S. J., Smit, B., Neaton, J. B., Long, J. R., & Brown, C. M. (2014). Comprehensive study of carbon dioxide adsorption in the metal–organic frameworks M2(dobdc) (M = Mg, Mn, Fe, Co, Ni, Cu, Zn). Chem. Sci., 5(12), 4569–4581. 10.1039/c4sc02064b
- Yu, D., Yazaydin, A. O., Lane, J. R., Dietzel, P. D. C., & Snurr, R. Q. (2013). A combined experimental and quantum chemical study of CO2 adsorption in the metal–organic framework CPO-27 with different metals. Chemical Science, 4(9), 3544. 10.1039/c3sc51319j
- Alonso, G., Bahamon, D., Keshavarz, F., Giménez, X., Gamallo, P., & Sayós, R. (2018). Density Functional Theory-Based Adsorption Isotherms for Pure and Flue Gas Mixtures on Mg-MOF-74. Application in CO2 Capture Swing Adsorption Processes. The Journal of Physical Chemistry C, 122(7), 3945–3957. 10.1021/acs.jpcc.8b00938
- Witman, M., Ling, S., Jawahery, S., Boyd, P. G., Haranczyk, M., Slater, B., & Smit, B. (2017). The Influence of Intrinsic Framework Flexibility on Adsorption in Nanoporous Materials. Journal of the American Chemical Society, 139(15), 5547–5557. 10.1021/jacs.7b01688