This tutorial will walk you through a few examples of how you can use UMA. Each step is covered in more detail elsewhere in the documentation, but this is well suited to a ~1-2 hour tutorial session for researchers new to UMA but with some background in ASE and molecular simulations.
Before you start / installation¶
You need to get a HuggingFace account and request access to the UMA models.
You need a Huggingface account, request access to https://
Permissions: Read access to contents of all public gated repos you can access
Then, add the token as an environment variable (using huggingface-cli login:
# Enter token via huggingface-cli
! huggingface-cli loginor you can set the token via HF_TOKEN variable:
# Set token via env variable
import os
os.environ['HF_TOKEN'] = 'MYTOKEN'Installation process¶
It may be enough to use pip install fairchem-core. This gets you the latest version on PyPi (https://
Here we install some sub-packages. This can take 2-5 minutes to run.
! pip install fairchem-core fairchem-data-oc fairchem-applications-cattsunami x3dase# Check that packages are installed
!pip list | grep fairchemfairchem-applications-cattsunami 1.1.2.dev182+gbafec1089
fairchem-core 2.14.1.dev13+gbafec1089
fairchem-data-oc 1.0.3.dev182+gbafec1089
fairchem-data-omat 0.2.1.dev87+gbafec1089
import fairchem.core
fairchem.core.__version__'2.14.1.dev13+gbafec1089'Illustrative examples¶
These should just run, and are here to show some basic uses.
Spin gap energy - OMOL¶
This is the difference in energy between a triplet and single ground state for a CH2 radical. This downloads a ~1GB checkpoint the first time you run it.
We don’t set a device here, so we get a warning about using a CPU device. You can ignore that. If a CUDA environment is available, a GPU may be used to speed up the calculations.
from fairchem.core import FAIRChemCalculator, pretrained_mlip
predictor = pretrained_mlip.get_predict_unit("uma-s-1")WARNING:root:device was not explicitly set, using device='cuda'.
WARNING:root:If 'dataset_list' is provided in the config, the code assumes that each dataset maps to itself. Please use 'dataset_mapping' as 'dataset_list' is deprecated and will be removed in the future.
from ase.build import molecule
# singlet CH2
singlet = molecule("CH2_s1A1d")
singlet.info.update({"spin": 1, "charge": 0})
singlet.calc = FAIRChemCalculator(predictor, task_name="omol")
# triplet CH2
triplet = molecule("CH2_s3B1d")
triplet.info.update({"spin": 3, "charge": 0})
triplet.calc = FAIRChemCalculator(predictor, task_name="omol")
print(triplet.get_potential_energy() - singlet.get_potential_energy())-0.5508370399475098
Example of adsorbate relaxation - OC20¶
Here we just setup a Cu(100) slab with a CO on it and relax it.
We specify an explicit device in the predictor here, and avoid the warning.
from ase.build import add_adsorbate, fcc100, molecule
from ase.optimize import LBFGS
from fairchem.core import FAIRChemCalculator, pretrained_mlip
predictor = pretrained_mlip.get_predict_unit("uma-s-1")
calc = FAIRChemCalculator(predictor, task_name="oc20")
# Set up your system as an ASE atoms object
slab = fcc100("Cu", (3, 3, 3), vacuum=8, periodic=True)
adsorbate = molecule("CO")
add_adsorbate(slab, adsorbate, 2.0, "bridge")
slab.calc = calc
# Set up LBFGS dynamics object
opt = LBFGS(slab)
opt.run(0.05, 100)
print(slab.get_potential_energy())WARNING:root:device was not explicitly set, using device='cuda'.
WARNING:root:If 'dataset_list' is provided in the config, the code assumes that each dataset maps to itself. Please use 'dataset_mapping' as 'dataset_list' is deprecated and will be removed in the future.
Step Time Energy fmax
LBFGS: 0 22:37:22 -89.596203 11.451673
LBFGS: 1 22:37:22 -92.497567 6.543858
LBFGS: 2 22:37:22 -92.624425 7.536288
LBFGS: 3 22:37:22 -93.000906 3.715976
LBFGS: 4 22:37:23 -93.158685 3.479954
LBFGS: 5 22:37:23 -93.264148 2.256767
LBFGS: 6 22:37:23 -93.505247 1.133170
LBFGS: 7 22:37:23 -93.595935 0.991856
LBFGS: 8 22:37:23 -93.705357 0.683607
LBFGS: 9 22:37:23 -93.791548 0.506915
LBFGS: 10 22:37:23 -93.837929 0.363997
LBFGS: 11 22:37:23 -93.856807 0.349533
LBFGS: 12 22:37:23 -93.881771 0.498644
LBFGS: 13 22:37:23 -93.900196 0.432912
LBFGS: 14 22:37:23 -93.910015 0.156310
LBFGS: 15 22:37:24 -93.915885 0.170003
LBFGS: 16 22:37:24 -93.922146 0.211720
LBFGS: 17 22:37:24 -93.929014 0.260811
LBFGS: 18 22:37:24 -93.935162 0.183895
LBFGS: 19 22:37:24 -93.938071 0.057447
LBFGS: 20 22:37:24 -93.938499 0.039155
-93.93849899606094
Example bulk relaxation - OMAT¶
from ase.build import bulk
from ase.filters import FrechetCellFilter
from ase.optimize import FIRE
from fairchem.core import FAIRChemCalculator, pretrained_mlip
predictor = pretrained_mlip.get_predict_unit("uma-s-1")
calc = FAIRChemCalculator(predictor, task_name="omat")
atoms = bulk("Fe")
atoms.calc = calc
opt = FIRE(FrechetCellFilter(atoms))
opt.run(0.05, 100)
print(atoms.get_stress()) # !!!! We get stress now!WARNING:root:device was not explicitly set, using device='cuda'.
WARNING:root:If 'dataset_list' is provided in the config, the code assumes that each dataset maps to itself. Please use 'dataset_mapping' as 'dataset_list' is deprecated and will be removed in the future.
Step Time Energy fmax
FIRE: 0 22:37:27 -8.261158 0.651784
FIRE: 1 22:37:27 -8.271310 0.358119
FIRE: 2 22:37:27 -8.264588 1.650196
FIRE: 3 22:37:27 -8.273672 0.177966
FIRE: 4 22:37:27 -8.272634 0.269083
FIRE: 5 22:37:27 -8.272766 0.257552
FIRE: 6 22:37:27 -8.273009 0.234350
FIRE: 7 22:37:27 -8.273319 0.199201
FIRE: 8 22:37:28 -8.273635 0.151747
FIRE: 9 22:37:28 -8.273890 0.091455
FIRE: 10 22:37:28 -8.274015 0.017801
[1.5563025e-03 1.5562251e-03 1.5562990e-03 3.6033928e-08 1.4666438e-10
3.5133503e-08]
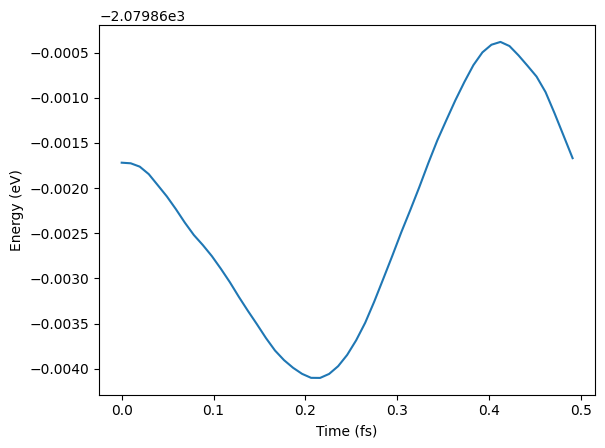
Molecular dynamics - OMOL¶
import matplotlib.pyplot as plt
from ase import units
from ase.build import molecule
from ase.io import Trajectory
from ase.md.langevin import Langevin
from fairchem.core import FAIRChemCalculator, pretrained_mlip
predictor = pretrained_mlip.get_predict_unit("uma-s-1")
calc = FAIRChemCalculator(predictor, task_name="omol")
atoms = molecule("H2O")
atoms.info.update(charge=0, spin=1) # For omol
atoms.calc = calc
dyn = Langevin(
atoms,
timestep=0.1 * units.fs,
temperature_K=400,
friction=0.001 / units.fs,
)
trajectory = Trajectory("my_md.traj", "w", atoms)
dyn.attach(trajectory.write, interval=1)
dyn.run(steps=50)
# See some results - not paper ready!
traj = Trajectory("my_md.traj")
plt.plot(
[i * 0.1 * units.fs for i in range(len(traj))],
[a.get_potential_energy() for a in traj],
)
plt.xlabel("Time (fs)")
plt.ylabel("Energy (eV)");WARNING:root:device was not explicitly set, using device='cuda'.
WARNING:root:If 'dataset_list' is provided in the config, the code assumes that each dataset maps to itself. Please use 'dataset_mapping' as 'dataset_list' is deprecated and will be removed in the future.
Catalyst Adsorption energies¶
The basic approach in computing an adsorption energy is to compute this energy difference:
dH = E_adslab - E_slab - E_adsWe use UMA for two of these energies E_adslab and E_slab. For E_ads We have to do something a little different. The OC20 task is not trained for molecules or molecular fragments. We use atomic energy reference energies instead. These are tabulated below.
The OC20 reference scheme is this reaction:
x CO + (x + y/2 - z) H2 + (z-x) H2O + w/2 N2 + * -> CxHyOzNw* For this example we have
-H2 + H2O + * -> O*. "O": -7.204 eVWhere "O": -7.204 is a constant.
To get the desired reaction energy we want we add the formation energy of water. We use either DFT or experimental values for this reaction energy.
1/2O2 + H2 -> H2OAlternatives to this approach are using DFT to estimate the energy of 1/2 O2, just make sure to use consistent settings with your task. You should not use OMOL for this.
from ase.build import add_adsorbate, fcc111
from ase.optimize import BFGS
from fairchem.core import FAIRChemCalculator, pretrained_mlip
predictor = pretrained_mlip.get_predict_unit("uma-s-1")
calc = FAIRChemCalculator(predictor, task_name="oc20")WARNING:root:device was not explicitly set, using device='cuda'.
WARNING:root:If 'dataset_list' is provided in the config, the code assumes that each dataset maps to itself. Please use 'dataset_mapping' as 'dataset_list' is deprecated and will be removed in the future.
# reference energies from a linear combination of H2O/N2/CO/H2!
atomic_reference_energies = {
"H": -3.477,
"N": -8.083,
"O": -7.204,
"C": -7.282,
}
re1 = -3.03 # Water formation energy from experiment
slab = fcc111("Pt", size=(2, 2, 5), vacuum=20.0)
slab.pbc = True
adslab = slab.copy()
add_adsorbate(adslab, "O", height=1.2, position="fcc")
slab.calc = calc
opt = BFGS(slab)
print("Relaxing slab")
opt.run(fmax=0.05, steps=100)
slab_e = slab.get_potential_energy()
adslab.calc = calc
opt = BFGS(adslab)
print("\nRelaxing adslab")
opt.run(fmax=0.05, steps=100)
adslab_e = adslab.get_potential_energy()Relaxing slab
Step Time Energy fmax
BFGS: 0 22:37:51 -104.710392 0.707696
BFGS: 1 22:37:51 -104.767890 0.605448
BFGS: 2 22:37:52 -104.919424 0.369264
BFGS: 3 22:37:52 -104.952876 0.441349
BFGS: 4 22:37:52 -105.030000 0.467593
BFGS: 5 22:37:52 -105.091450 0.365230
BFGS: 6 22:37:52 -105.128720 0.195037
BFGS: 7 22:37:52 -105.143315 0.048836
Relaxing adslab
Step Time Energy fmax
BFGS: 0 22:37:53 -110.055656 1.762239
BFGS: 1 22:37:53 -110.239039 0.996808
BFGS: 2 22:37:53 -110.389561 0.747533
BFGS: 3 22:37:53 -110.441195 0.818361
BFGS: 4 22:37:53 -110.557367 0.688425
BFGS: 5 22:37:54 -110.631217 0.497353
BFGS: 6 22:37:54 -110.687290 0.690802
BFGS: 7 22:37:54 -110.737890 0.729386
BFGS: 8 22:37:54 -110.774873 0.435721
BFGS: 9 22:37:55 -110.786666 0.199894
BFGS: 10 22:37:55 -110.789560 0.080700
BFGS: 11 22:37:55 -110.790038 0.057990
BFGS: 12 22:37:55 -110.790286 0.044006
Now we compute the adsorption energy.
# Energy for ((H2O-H2) + * -> *O) + (H2 + 1/2O2 -> H2O) leads to 1/2O2 + * -> *O!
adslab_e - slab_e - atomic_reference_energies["O"] + re1-1.4729707438775743How did we do? We need a reference point. In the paper below, there is an atomic adsorption energy for O on Pt(111) of about -4.264 eV. This is for the reaction O + * -> O*. To convert this to the dissociative adsorption energy, we have to add the reaction:
1/2 O2 -> O D = 2.58 eV (expt)to get a comparable energy of about -1.68 eV. There is about ~0.2 eV difference (we predicted -1.47 eV above, and the reference comparison is -1.68 eV) to account for. The biggest difference is likely due to the differences in exchange-correlation functional. The reference data used the PBE functional, and eSCN was trained on RPBE data. To additional places where there are differences include:
Difference in lattice constant
The reference energy used for the experiment references. These can differ by up to 0.5 eV from comparable DFT calculations.
How many layers are relaxed in the calculation
Some of these differences tend to be systematic, and you can calibrate and correct these, especially if you can augment these with your own DFT calculations.
It is always a good idea to visualize the geometries to make sure they look reasonable.
import matplotlib.pyplot as plt
from ase.visualize.plot import plot_atoms
fig, axs = plt.subplots(1, 2)
plot_atoms(slab, axs[0])
plot_atoms(slab, axs[1], rotation=("-90x"))
axs[0].set_axis_off()
axs[1].set_axis_off()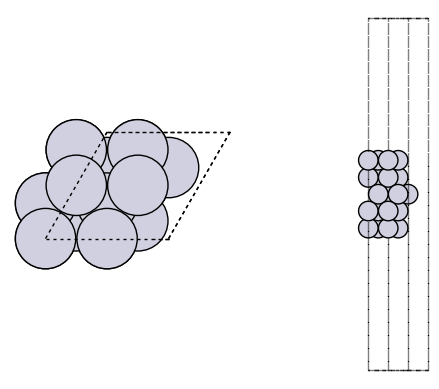
fig, axs = plt.subplots(1, 2)
plot_atoms(adslab, axs[0])
plot_atoms(adslab, axs[1], rotation=("-90x"))
axs[0].set_axis_off()
axs[1].set_axis_off()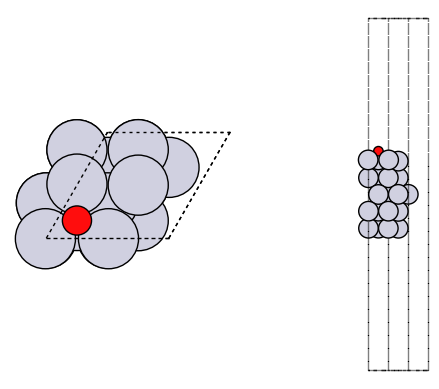
Molecular vibrations¶
from ase import Atoms
from ase.optimize import BFGS
predictor = pretrained_mlip.get_predict_unit("uma-s-1")
calc = FAIRChemCalculator(predictor, task_name="omol")
from ase.vibrations import Vibrations
n2 = Atoms("N2", [(0, 0, 0), (0, 0, 1.1)])
n2.info.update({"spin": 1, "charge": 0})
n2.calc = calc
BFGS(n2).run(fmax=0.01)WARNING:root:device was not explicitly set, using device='cuda'.
WARNING:root:If 'dataset_list' is provided in the config, the code assumes that each dataset maps to itself. Please use 'dataset_mapping' as 'dataset_list' is deprecated and will be removed in the future.
Step Time Energy fmax
BFGS: 0 22:38:09 -2981.068009 1.645286
BFGS: 1 22:38:09 -2980.961841 6.601526
BFGS: 2 22:38:09 -2981.076753 0.203644
BFGS: 3 22:38:09 -2981.076882 0.024168
BFGS: 4 22:38:09 -2981.076883 0.000103
np.True_vib = Vibrations(n2)
vib.run()
vib.summary()---------------------
# meV cm^-1
---------------------
0 0.0 0.0
1 0.0 0.0
2 0.0 0.0
3 2.0 16.1
4 2.0 16.1
5 309.2 2494.2
---------------------
Zero-point energy: 0.157 eV
Bulk alloy phase behavior¶
Adapted from https://
We manually compute the formation energy of pure compounds and some alloy compositions to assess stability.
from ase.atoms import Atom, Atoms
from ase.filters import FrechetCellFilter
from ase.optimize import FIRE
from fairchem.core import FAIRChemCalculator, pretrained_mlip
predictor = pretrained_mlip.get_predict_unit("uma-s-1")
cu = Atoms(
[Atom("Cu", [0.000, 0.000, 0.000])],
cell=[[1.818, 0.000, 1.818], [1.818, 1.818, 0.000], [0.000, 1.818, 1.818]],
pbc=True,
)
cu.calc = FAIRChemCalculator(predictor, task_name="omat")
opt = FIRE(FrechetCellFilter(cu))
opt.run(0.05, 100)
cu.get_potential_energy()WARNING:root:device was not explicitly set, using device='cuda'.
WARNING:root:If 'dataset_list' is provided in the config, the code assumes that each dataset maps to itself. Please use 'dataset_mapping' as 'dataset_list' is deprecated and will be removed in the future.
Step Time Energy fmax
FIRE: 0 22:38:15 -3.756933 0.161999
FIRE: 1 22:38:15 -3.757594 0.110083
FIRE: 2 22:38:15 -3.758130 0.020767
-3.7581299773902463pd = Atoms(
[Atom("Pd", [0.000, 0.000, 0.000])],
cell=[[1.978, 0.000, 1.978], [1.978, 1.978, 0.000], [0.000, 1.978, 1.978]],
pbc=True,
)
pd.calc = FAIRChemCalculator(predictor, task_name="omat")
opt = FIRE(FrechetCellFilter(pd))
opt.run(0.05, 100)
pd.get_potential_energy() Step Time Energy fmax
FIRE: 0 22:38:15 -5.211726 0.240058
FIRE: 1 22:38:15 -5.213070 0.131579
FIRE: 2 22:38:16 -5.213503 0.060259
FIRE: 3 22:38:16 -5.213528 0.051645
FIRE: 4 22:38:16 -5.213565 0.035871
-5.213564622137426Alloy formation energies¶
cupd1 = Atoms(
[Atom("Cu", [0.000, 0.000, 0.000]), Atom("Pd", [-1.652, 0.000, 2.039])],
cell=[[0.000, -2.039, 2.039], [0.000, 2.039, 2.039], [-3.303, 0.000, 0.000]],
pbc=True,
) # Note pbc=True is important, it is not the default and OMAT
cupd1.calc = FAIRChemCalculator(predictor, task_name="omat")
opt = FIRE(FrechetCellFilter(cupd1))
opt.run(0.05, 100)
cupd1.get_potential_energy() Step Time Energy fmax
FIRE: 0 22:38:16 -9.202821 0.142029
FIRE: 1 22:38:17 -9.203042 0.127498
FIRE: 2 22:38:17 -9.203371 0.101174
FIRE: 3 22:38:17 -9.203669 0.068562
FIRE: 4 22:38:17 -9.203892 0.060712
FIRE: 5 22:38:17 -9.204129 0.078850
FIRE: 6 22:38:18 -9.204490 0.081598
FIRE: 7 22:38:18 -9.204987 0.069264
FIRE: 8 22:38:18 -9.205592 0.045644
-9.205591734189348cupd2 = Atoms(
[
Atom("Cu", [-0.049, 0.049, 0.049]),
Atom("Cu", [-11.170, 11.170, 11.170]),
Atom("Pd", [-7.415, 7.415, 7.415]),
Atom("Pd", [-3.804, 3.804, 3.804]),
],
cell=[[-5.629, 3.701, 5.629], [-3.701, 5.629, 5.629], [-5.629, 5.629, 3.701]],
pbc=True,
)
cupd2.calc = FAIRChemCalculator(predictor, task_name="omat")
opt = FIRE(FrechetCellFilter(cupd2))
opt.run(0.05, 100)
cupd2.get_potential_energy() Step Time Energy fmax
FIRE: 0 22:38:18 -18.126594 0.181633
FIRE: 1 22:38:18 -18.127546 0.162952
FIRE: 2 22:38:19 -18.129066 0.127293
FIRE: 3 22:38:19 -18.130534 0.078068
FIRE: 4 22:38:19 -18.131346 0.021485
-18.13134633687845# Delta Hf cupd-1 = -0.11 eV/atom
hf1 = (
cupd1.get_potential_energy() - cu.get_potential_energy() - pd.get_potential_energy()
)
hf1-0.2338971346616754# DFT: Delta Hf cupd-2 = -0.04 eV/atom
hf2 = (
cupd2.get_potential_energy()
- 2 * cu.get_potential_energy()
- 2 * pd.get_potential_energy()
)
hf2-0.18795713782310663hf1 - hf2, (-0.11 - -0.04)(-0.04593999683856875, -0.07)These indicate that cupd-1 and cupd-2 are both more stable than phase separated Cu and Pd, and that cupd-1 is more stable than cupd-2. The absolute formation energies differ from the DFT references, but the relative differences are quite close. The absolute differences could be due to DFT parameter choices (XC, psp, etc.).
Phonon calculation¶
This takes 4-10 minutes. Adapted from https://
Phonons have applications in computing the stability and free energy of solids. See:
https://
www .sciencedirect .com /science /article /pii /S1359646215003127 https://
iopscience .iop .org /book /mono /978 -0 -7503 -2572 -1 /chapter /bk978 -0 -7503 -2572 -1ch1
from ase.build import bulk
from ase.phonons import Phonons
predictor = pretrained_mlip.get_predict_unit("uma-s-1")
calc = FAIRChemCalculator(predictor, task_name="omat")
# Setup crystal
atoms = bulk("Al", "fcc", a=4.05)
# Phonon calculator
N = 7
ph = Phonons(atoms, calc, supercell=(N, N, N), delta=0.05)
ph.run()
# Read forces and assemble the dynamical matrix
ph.read(acoustic=True)
ph.clean()
path = atoms.cell.bandpath("GXULGK", npoints=100)
bs = ph.get_band_structure(path)
dos = ph.get_dos(kpts=(20, 20, 20)).sample_grid(npts=100, width=1e-3)WARNING:root:device was not explicitly set, using device='cuda'.
WARNING:root:If 'dataset_list' is provided in the config, the code assumes that each dataset maps to itself. Please use 'dataset_mapping' as 'dataset_list' is deprecated and will be removed in the future.
WARNING, 1 imaginary frequencies at q = ( 0.00, 0.00, 0.00) ; (omega_q = 6.562e-09*i)
WARNING, 1 imaginary frequencies at q = ( 0.00, 0.00, 0.00) ; (omega_q = 6.562e-09*i)
# Plot the band structure and DOS:
import matplotlib.pyplot as plt # noqa
fig = plt.figure(figsize=(7, 4))
ax = fig.add_axes([0.12, 0.07, 0.67, 0.85])
emax = 0.04
bs.plot(ax=ax, emin=0.0, emax=emax)
dosax = fig.add_axes([0.8, 0.07, 0.17, 0.85])
dosax.fill_between(
dos.get_weights(),
dos.get_energies(),
y2=0,
color="grey",
edgecolor="k",
lw=1,
)
dosax.set_ylim(0, emax)
dosax.set_yticks([])
dosax.set_xticks([])
dosax.set_xlabel("DOS", fontsize=18);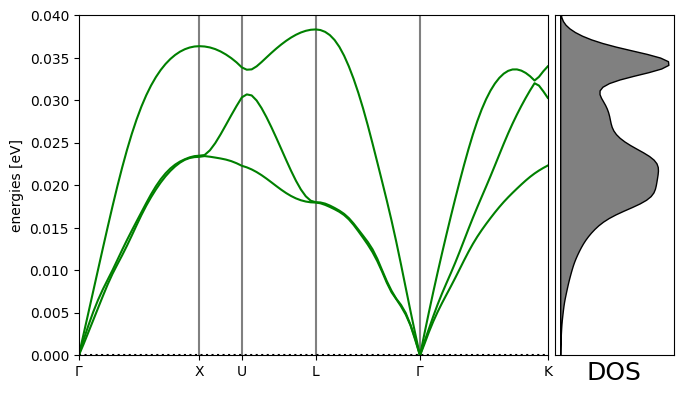
Transition States (NEBs)¶
Nudged elastic band calculations are among the most costly calculations we do. UMA makes these quicker!
We explore diffusion of an O adatom from an hcp to an fcc site on Pt(111).
Initial state¶
from ase.build import add_adsorbate, fcc111, molecule
from ase.optimize import LBFGS
from fairchem.core import FAIRChemCalculator, pretrained_mlip
predictor = pretrained_mlip.get_predict_unit("uma-s-1")
calc = FAIRChemCalculator(predictor, task_name="oc20")
# Set up your system as an ASE atoms object
initial = fcc111("Pt", (3, 3, 3), vacuum=8, periodic=True)
adsorbate = molecule("O")
add_adsorbate(initial, adsorbate, 2.0, "fcc")
initial.calc = calc
# Set up LBFGS dynamics object
opt = LBFGS(initial)
opt.run(0.05, 100)
print(initial.get_potential_energy())WARNING:root:device was not explicitly set, using device='cuda'.
WARNING:root:If 'dataset_list' is provided in the config, the code assumes that each dataset maps to itself. Please use 'dataset_mapping' as 'dataset_list' is deprecated and will be removed in the future.
Step Time Energy fmax
LBFGS: 0 22:38:32 -141.329799 3.509945
LBFGS: 1 22:38:32 -141.719926 3.515035
LBFGS: 2 22:38:32 -142.980944 2.978369
LBFGS: 3 22:38:32 -143.684058 0.968201
LBFGS: 4 22:38:33 -143.787051 1.271678
LBFGS: 5 22:38:33 -143.858780 0.874625
LBFGS: 6 22:38:33 -143.933926 0.170912
LBFGS: 7 22:38:33 -143.937118 0.152467
LBFGS: 8 22:38:33 -143.944594 0.122230
LBFGS: 9 22:38:33 -143.948826 0.109292
LBFGS: 10 22:38:33 -143.952235 0.069972
LBFGS: 11 22:38:33 -143.953715 0.080109
LBFGS: 12 22:38:33 -143.955176 0.083486
LBFGS: 13 22:38:34 -143.956798 0.066267
LBFGS: 14 22:38:34 -143.958312 0.031494
-143.9583123027719
Final state¶
# Set up your system as an ASE atoms object
final = fcc111("Pt", (3, 3, 3), vacuum=8, periodic=True)
adsorbate = molecule("O")
add_adsorbate(final, adsorbate, 2.0, "hcp")
final.calc = FAIRChemCalculator(predictor, task_name="oc20")
# Set up LBFGS dynamics object
opt = LBFGS(final)
opt.run(0.05, 100)
print(final.get_potential_energy()) Step Time Energy fmax
LBFGS: 0 22:38:34 -141.282604 3.340431
LBFGS: 1 22:38:34 -141.659791 3.323329
LBFGS: 2 22:38:34 -142.891403 2.596618
LBFGS: 3 22:38:34 -143.418900 1.225932
LBFGS: 4 22:38:34 -143.484096 0.977172
LBFGS: 5 22:38:34 -143.606344 0.136688
LBFGS: 6 22:38:34 -143.610842 0.118672
LBFGS: 7 22:38:34 -143.613319 0.100140
LBFGS: 8 22:38:34 -143.615008 0.078482
LBFGS: 9 22:38:35 -143.616456 0.051598
LBFGS: 10 22:38:35 -143.617143 0.033145
-143.6171429454721
Setup and relax the band¶
from ase.mep import NEB
images = [initial]
for i in range(3):
image = initial.copy()
image.calc = FAIRChemCalculator(predictor, task_name="oc20")
images.append(image)
images.append(final)
neb = NEB(images)
neb.interpolate()
opt = LBFGS(neb, trajectory="neb.traj")
opt.run(0.05, 100) Step Time Energy fmax
LBFGS: 0 22:38:35 -143.193976 3.039304
LBFGS: 1 22:38:35 -143.360199 1.460890
LBFGS: 2 22:38:36 -143.411349 0.450311
LBFGS: 3 22:38:36 -143.423401 0.447556
LBFGS: 4 22:38:36 -143.443053 0.476386
LBFGS: 5 22:38:36 -143.459314 0.378509
LBFGS: 6 22:38:37 -143.469904 0.211695
LBFGS: 7 22:38:37 -143.474784 0.177272
LBFGS: 8 22:38:37 -143.475807 0.183705
LBFGS: 9 22:38:38 -143.477288 0.178492
LBFGS: 10 22:38:38 -143.478786 0.167194
LBFGS: 11 22:38:38 -143.479388 0.094900
LBFGS: 12 22:38:38 -143.479539 0.096950
LBFGS: 13 22:38:39 -143.479790 0.100285
LBFGS: 14 22:38:39 -143.480345 0.124778
LBFGS: 15 22:38:39 -143.481008 0.092787
LBFGS: 16 22:38:40 -143.481435 0.050531
LBFGS: 17 22:38:40 -143.481732 0.040589
np.True_from ase.mep import NEBTools
NEBTools(neb.images).plot_band();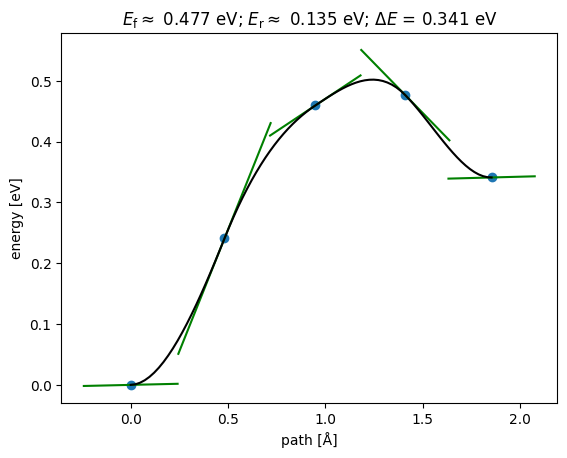
This could be a good initial guess to initialize an NEB in DFT.
Ideas for things you can do with UMA¶
Advanced applications¶
These take a while to run.
AdsorbML¶
It is so cheap to run these calculations that we can screen a broad range of adsorbate sites and rank them in stability. The AdsorbML approach automates this. This takes quite a while to run here, and we don’t do it in the workshop.
Expert adsorption energies¶
This tutorial reproduces Fig 6b from the following paper: Zhou, Jing, et al. “Enhanced Catalytic Activity of Bimetallic Ordered Catalysts for Nitrogen Reduction Reaction by Perturbation of Scaling Relations.” ACS Catalysis 134 (2023): 2190-2201 (Zhou et al. (2023)).
This takes up to an hour with a GPU, and much longer with a CPU.
CatTsunami¶
The CatTsunami tutorial is an example of enumerating initial and final states, and computing reaction paths between them with UMA.
Acknowledgments¶
This tutorial was originally compiled by John Kitchin (CMU) for the NAM29 catalysis tutorial session, using a variety of resources from the FAIR chemistry repository.
- Musielewicz, J., Wang, X., Tian, T., & Ulissi, Z. (2022). FINETUNA: fine-tuning accelerated molecular simulations. Machine Learning: Science and Technology, 3(3), 03LT01. 10.1088/2632-2153/ac8fe0
- Wang, X., Musielewicz, J., Tran, R., Kumar Ethirajan, S., Fu, X., Mera, H., Kitchin, J. R., Kurchin, R. C., & Ulissi, Z. W. (2024). Generalization of graph-based active learning relaxation strategies across materials. Machine Learning: Science and Technology, 5(2), 025018. 10.1088/2632-2153/ad37f0
- Wander, B., Musielewicz, J., Cheula, R., & Kitchin, J. R. (2025). Accessing Numerical Energy Hessians with Graph Neural Network Potentials and Their Application in Heterogeneous Catalysis. The Journal of Physical Chemistry C, 129(7), 3510–3521. 10.1021/acs.jpcc.4c07477
- Zhou, J., Chen, X., Guo, M., Hu, W., Huang, B., & Yuan, D. (2023). Enhanced Catalytic Activity of Bimetallic Ordered Catalysts for Nitrogen Reduction Reaction by Perturbation of Scaling Relations. ACS Catalysis, 13(4), 2190–2201. 10.1021/acscatal.2c05877